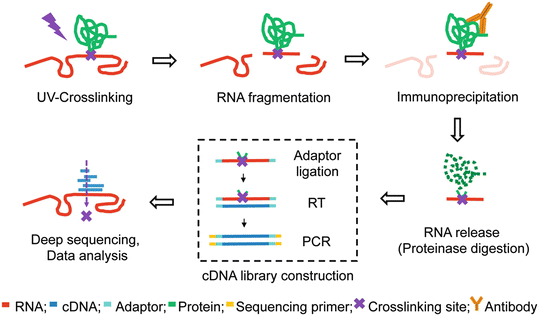
Outline of HITS-CLIP, PAR-CLIP and several variants, iCLIP, iCLAP and... | Download Scientific Diagram

Puf5p HITS-CLIP. (a) Summary of HITS-CLIP protocol. S. cerevisiae cells... | Download Scientific Diagram

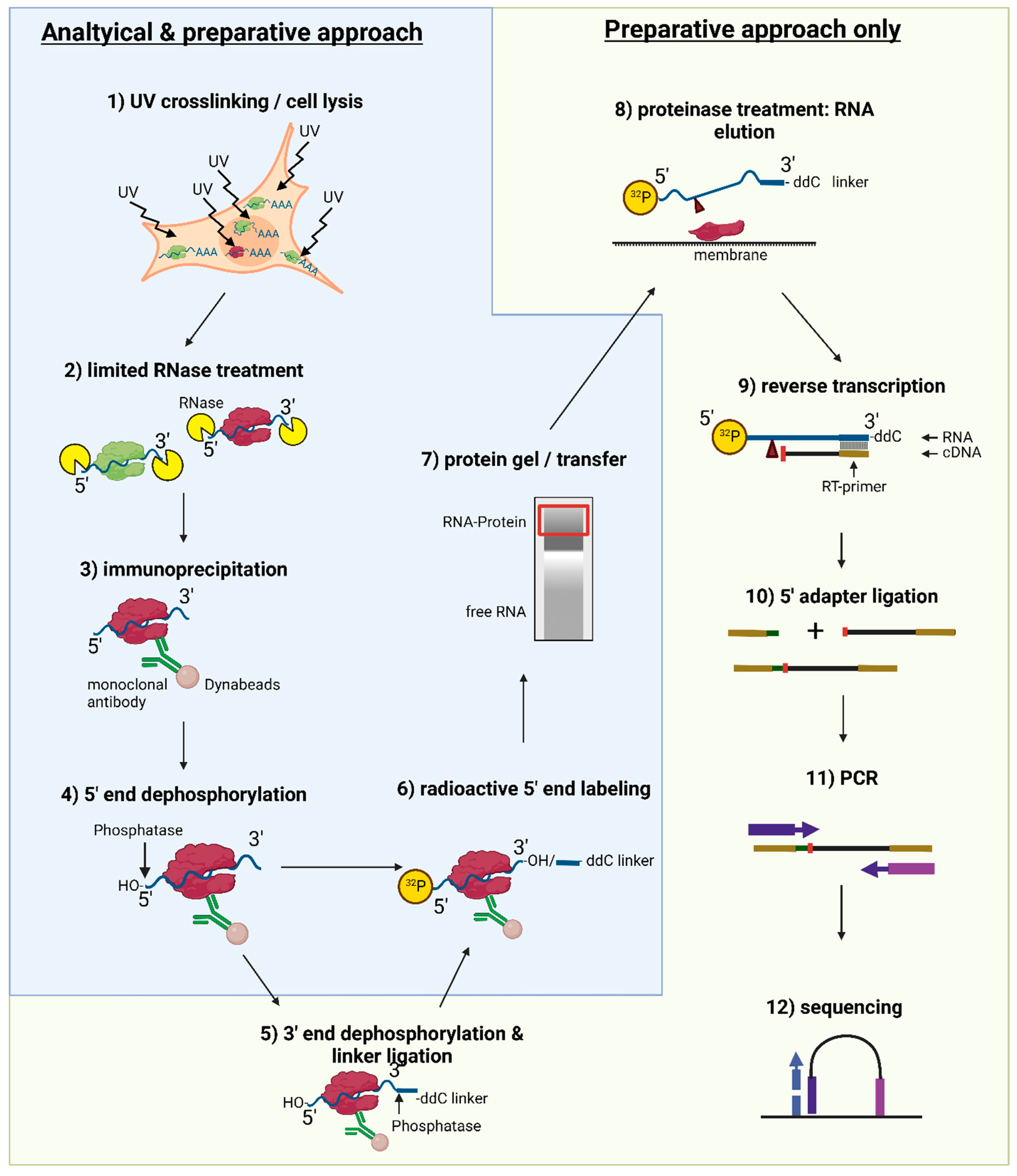
iCLIP - Transcriptome-wide Mapping of Protein-RNA Interactions with Individual Nucleotide Resolution | Protocol

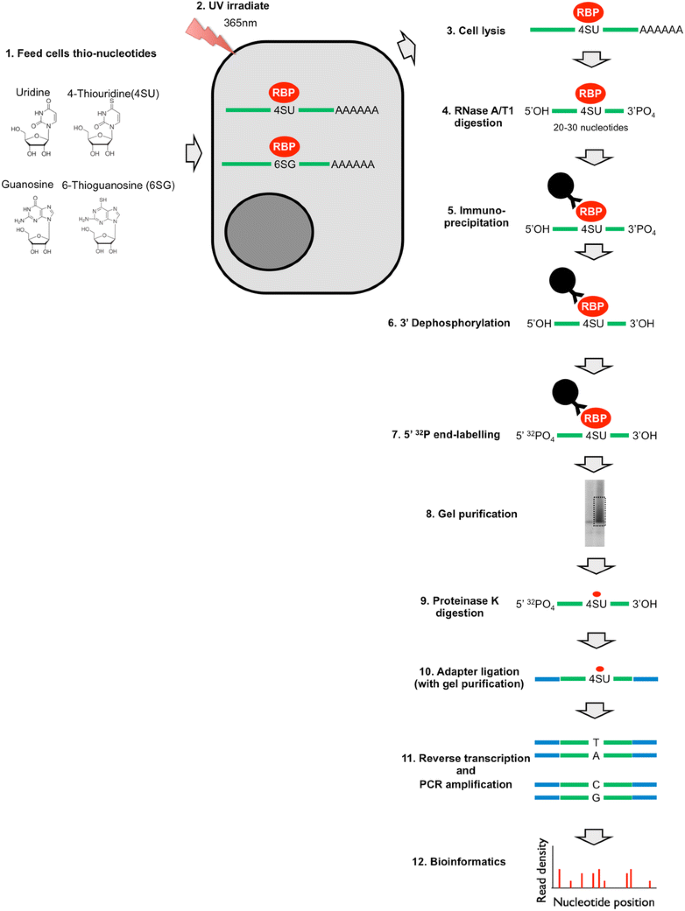
PAR-CLIP and Streamlined Small RNA cDNA Library Preparation Protocol for the Identification of RNA Binding Protein Target Sites | RNA-Seq Blog
MPs | Free Full-Text | Studying miRNA–mRNA Interactions: An Optimized CLIP-Protocol for Endogenous Ago2-Protein
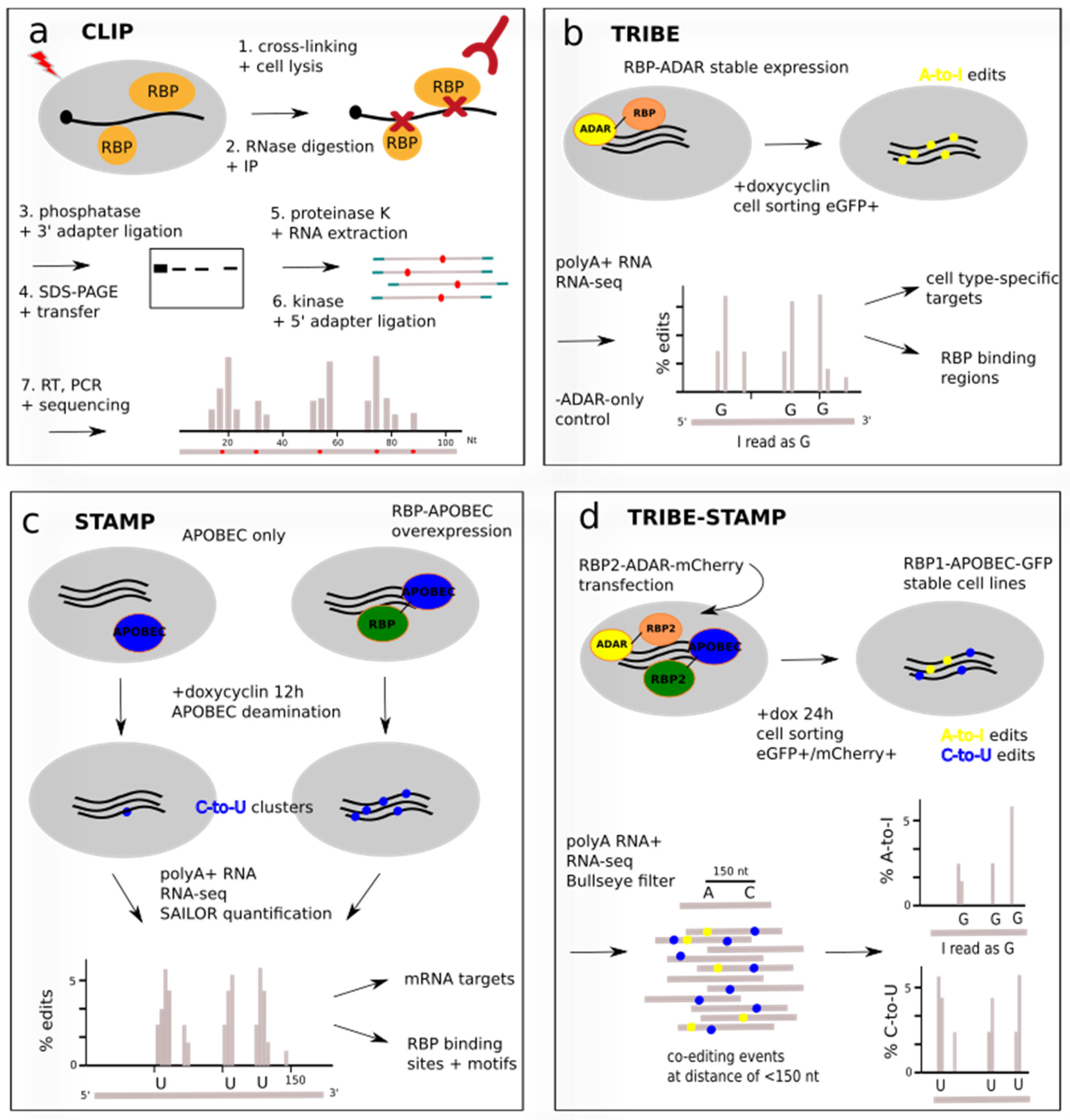
BioChem | Free Full-Text | Current Technical Approaches to Study RNA–Protein Interactions in mRNAs and Long Non-Coding RNAs

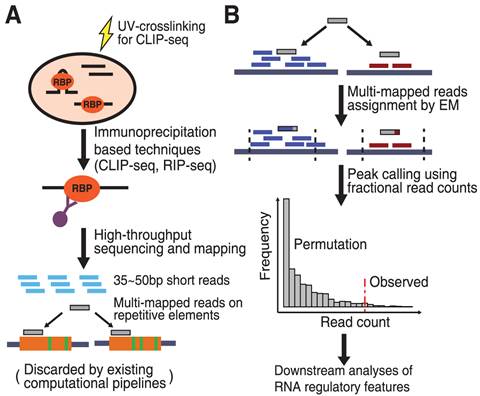
ACE-score-based Analysis of Temporal miRNA Targetomes During Human Cytomegalovirus Infection Using AGO-CLIP-seq
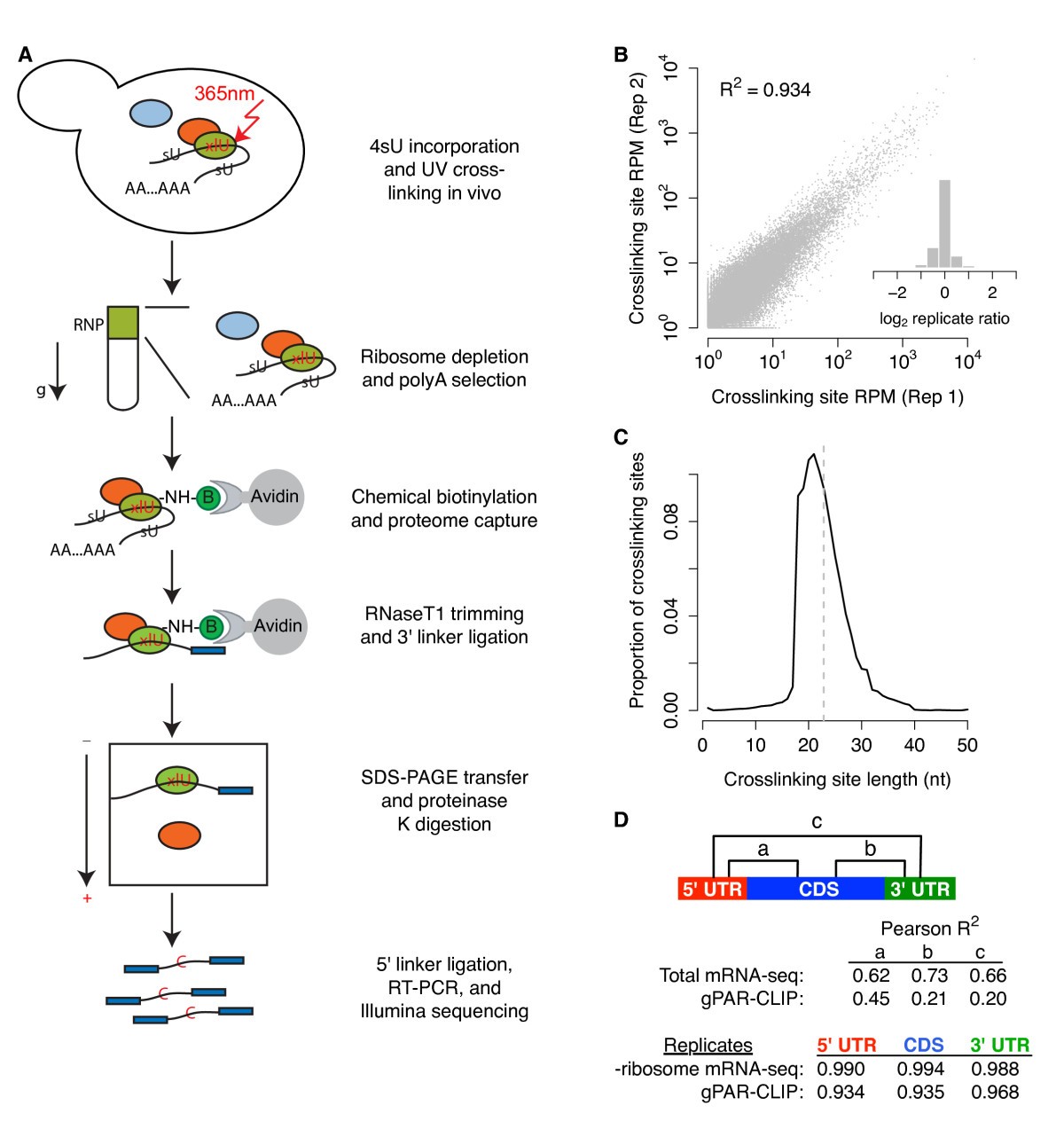
Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae | Genome Biology | Full Text
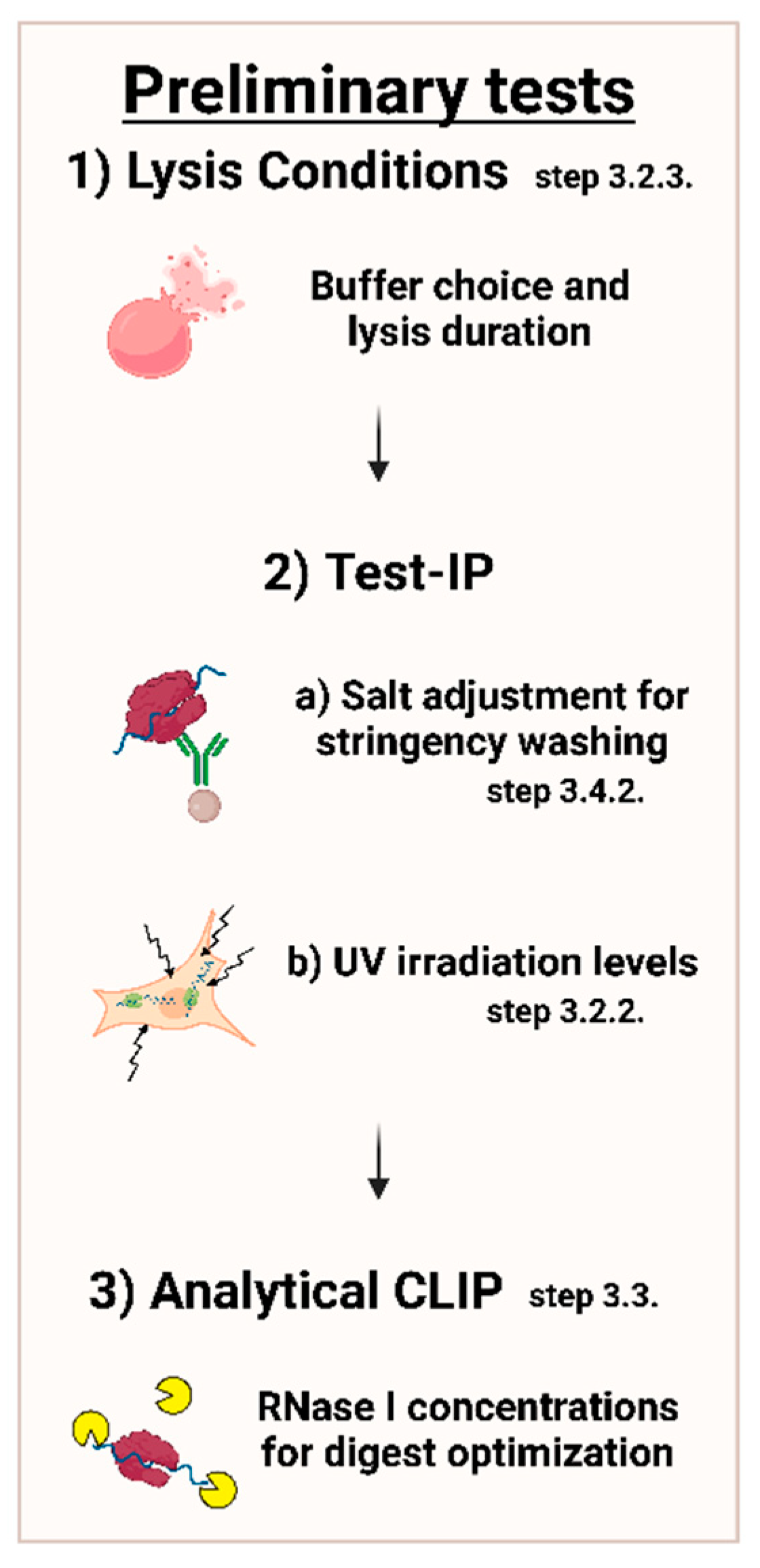
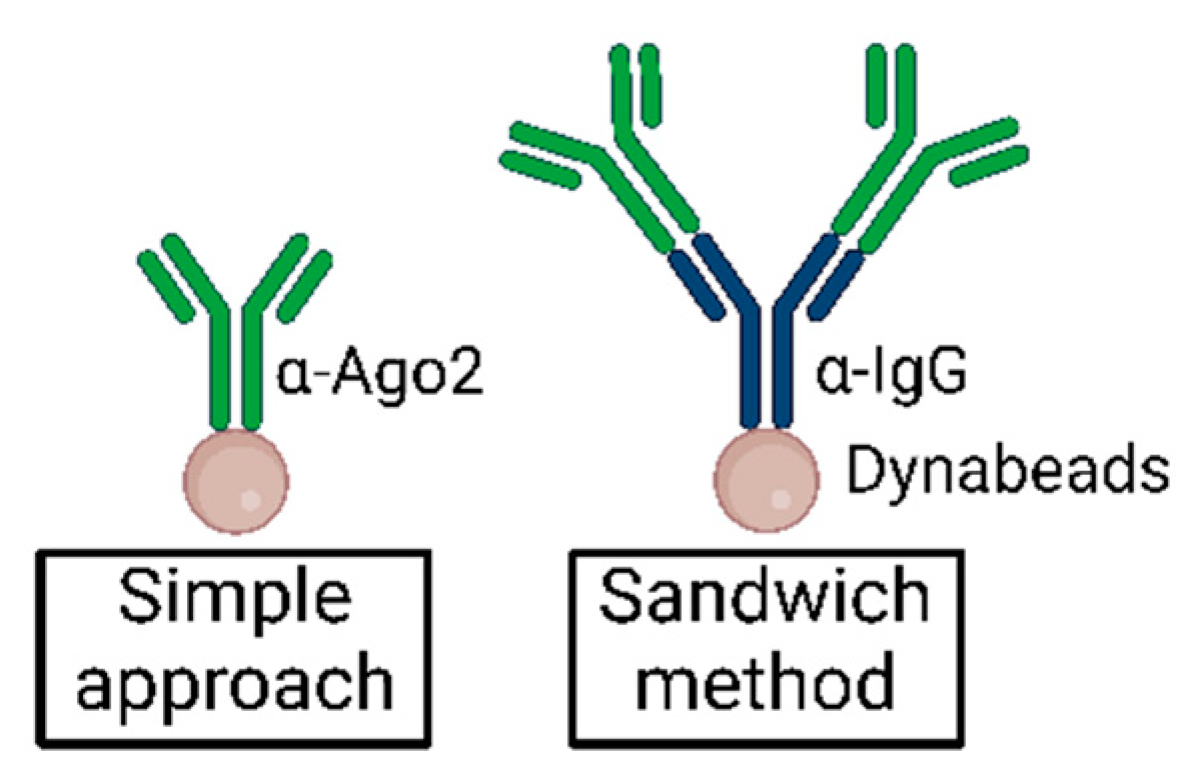
MPs | Free Full-Text | Studying miRNA–mRNA Interactions: An Optimized CLIP-Protocol for Endogenous Ago2-Protein
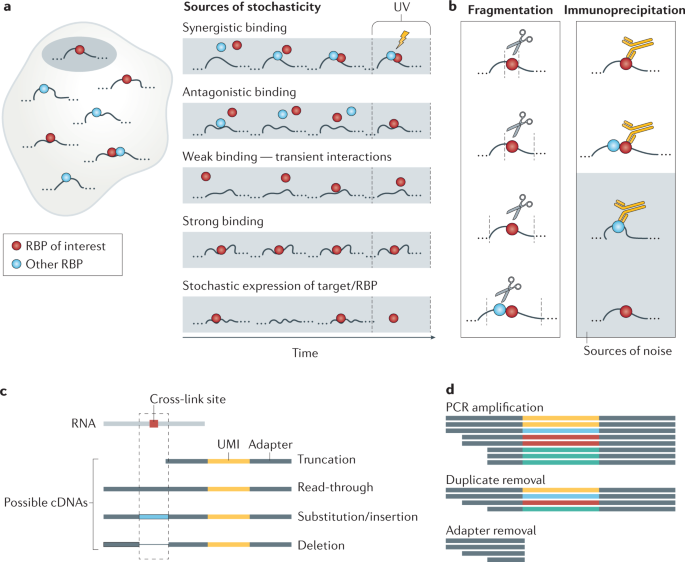




-1.jpg)